Effect of Protein Corona on The Transfection Efficiency of Lipid-Coated Graphene Oxide-Based Cell Transfection Reagents
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of GOCL Nanoparticles
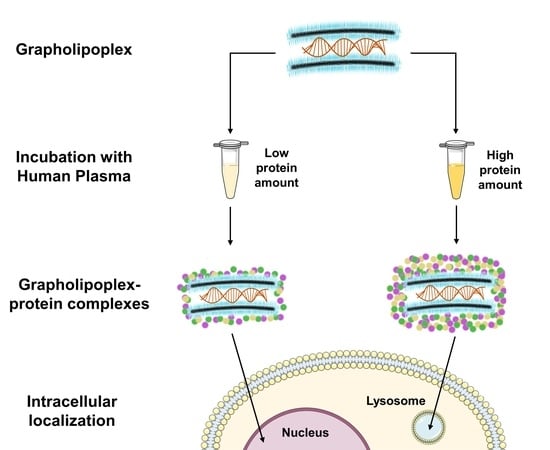
2.3. Preparation of Grapholipoplexes
2.4. Preparation of Grapholipopex-Protein Complexes
2.5. Size and Zeta–Potential Measurements
2.6. Cell Culture
2.7. Transfection Efficiency Experiments
2.8. Cell Viability Experiments
2.9. 1D SDS PAGE Gel Electrophoresis
2.10. Confocal Microscopy
2.11. Statistical Analysis
3. Results
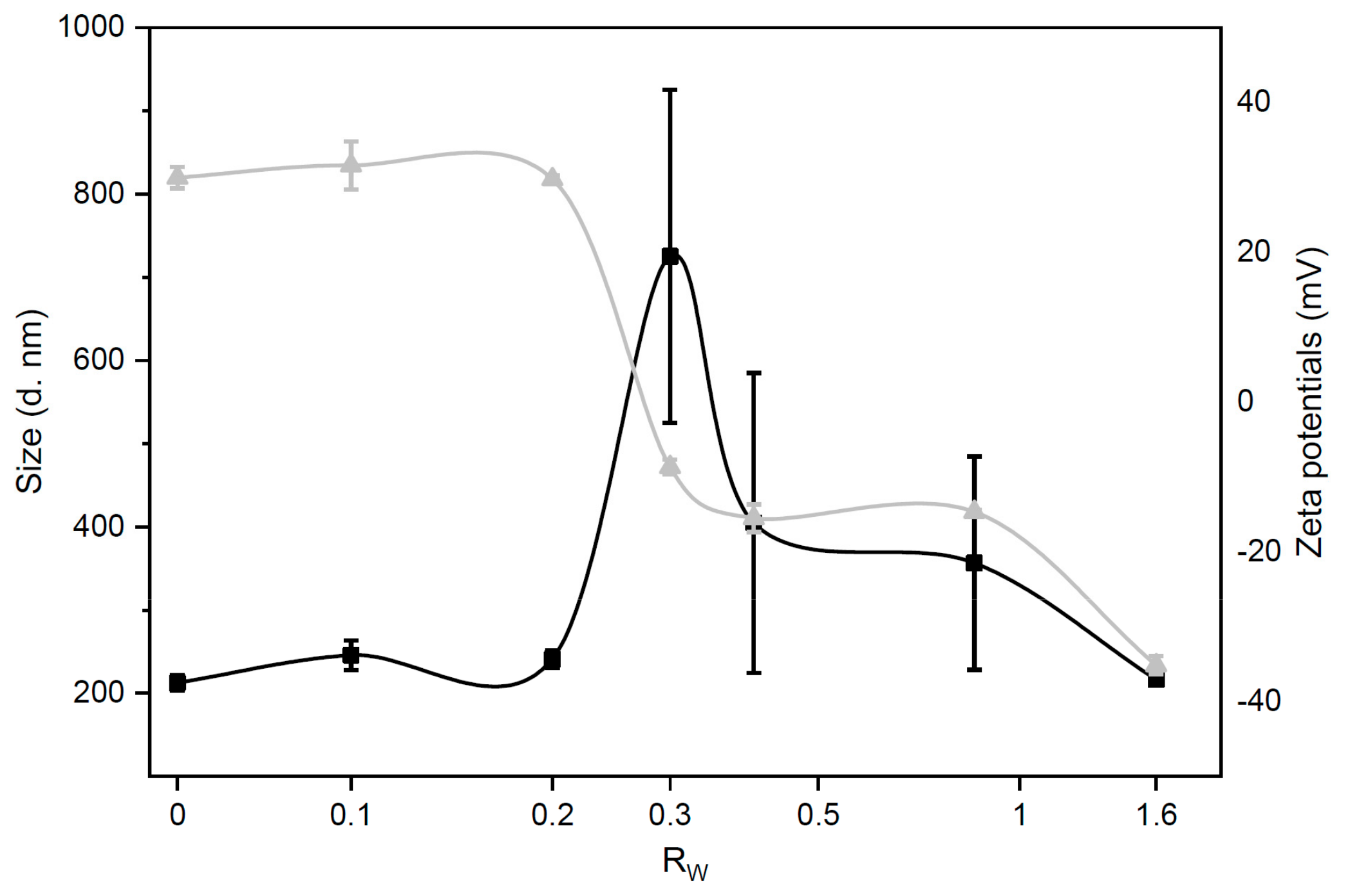
3.1. Size and Zeta-Potential of Grapholipoplexes
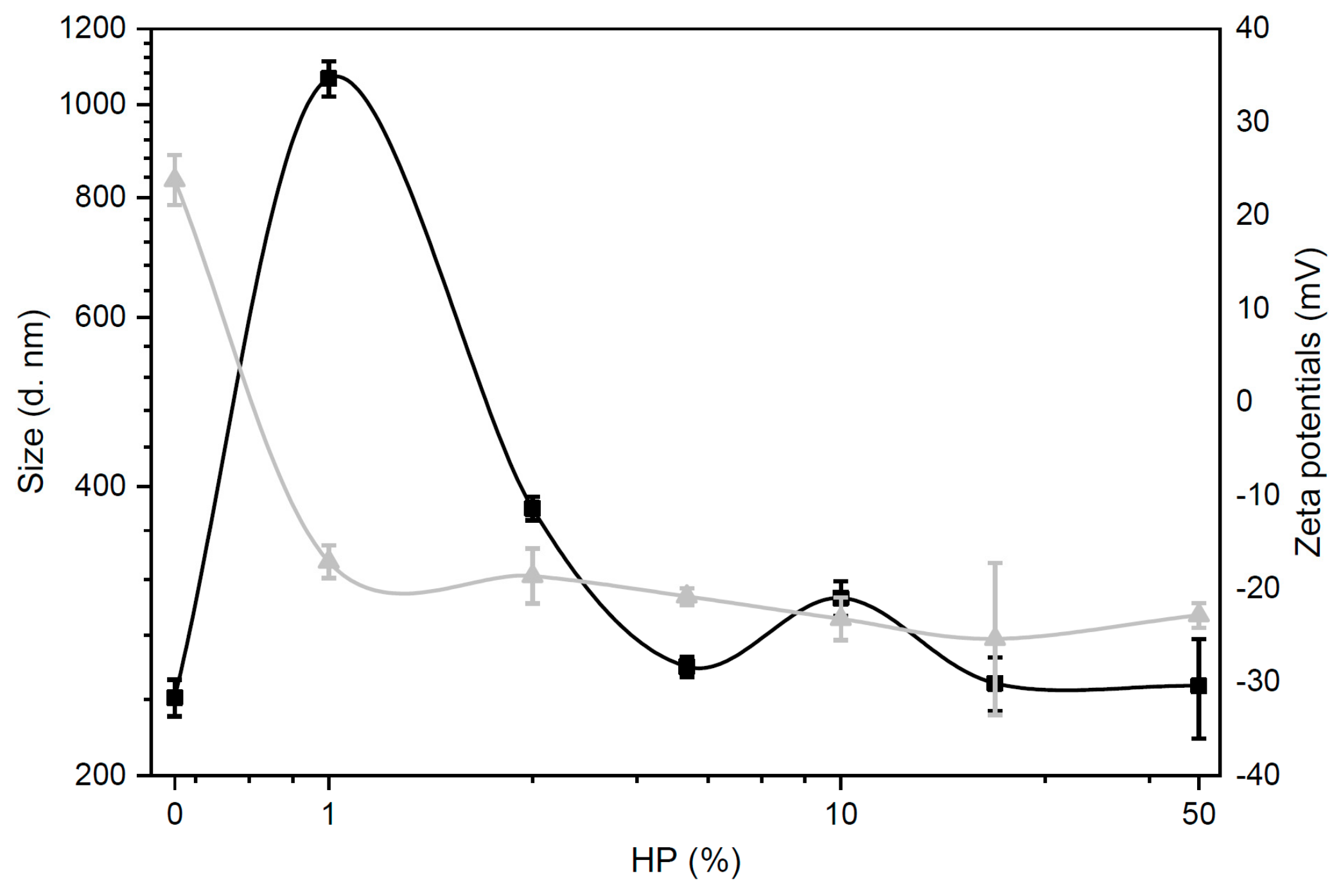
3.2. Size and Zeta-Potential of Biocoronated Grapholipoplexes
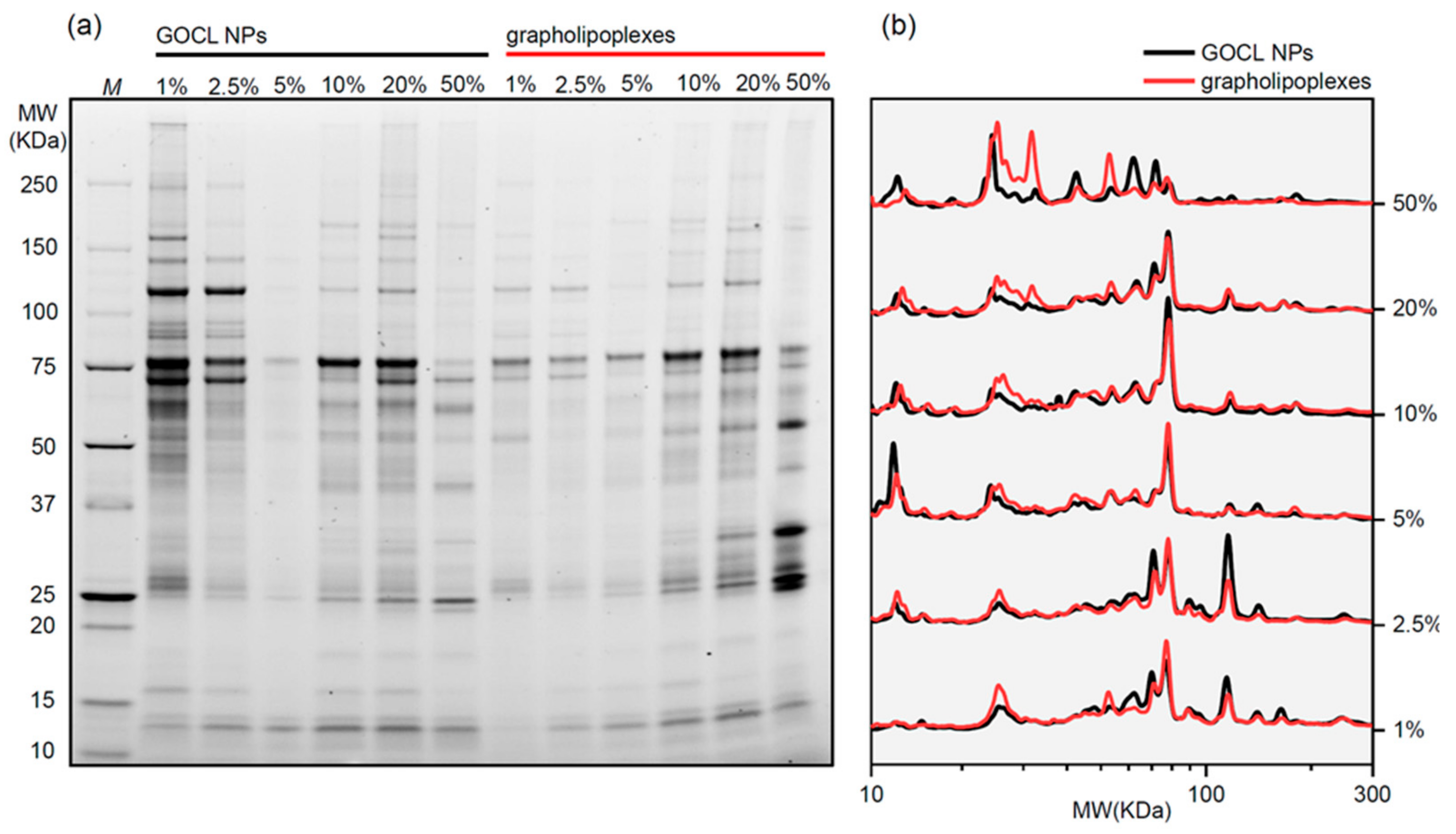
3.3. Protein Corona of Grapholipoplexes
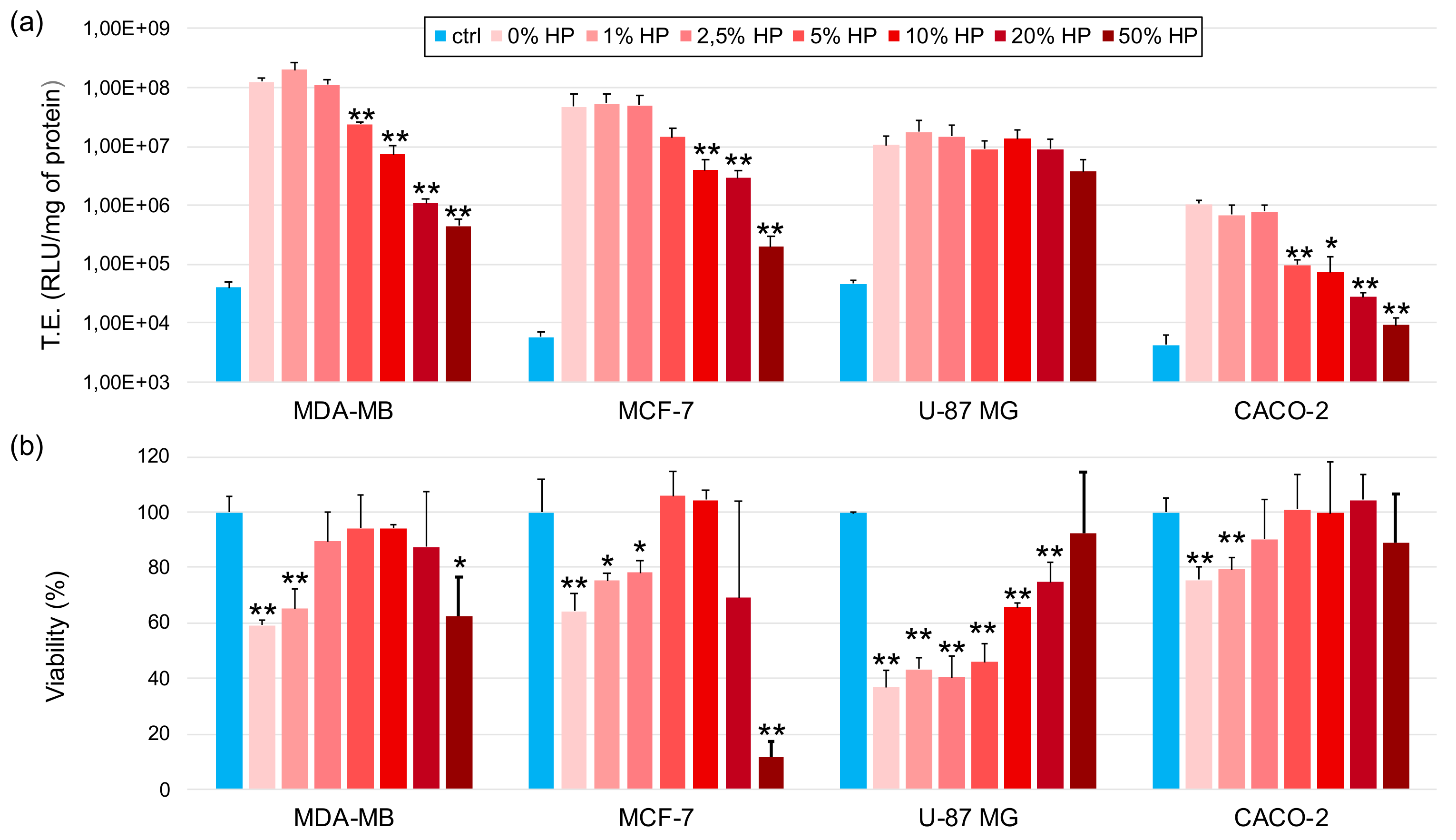
3.4. Transfection Efficiency and Cytotoxicity of Biocorononated Grapholipoplexes
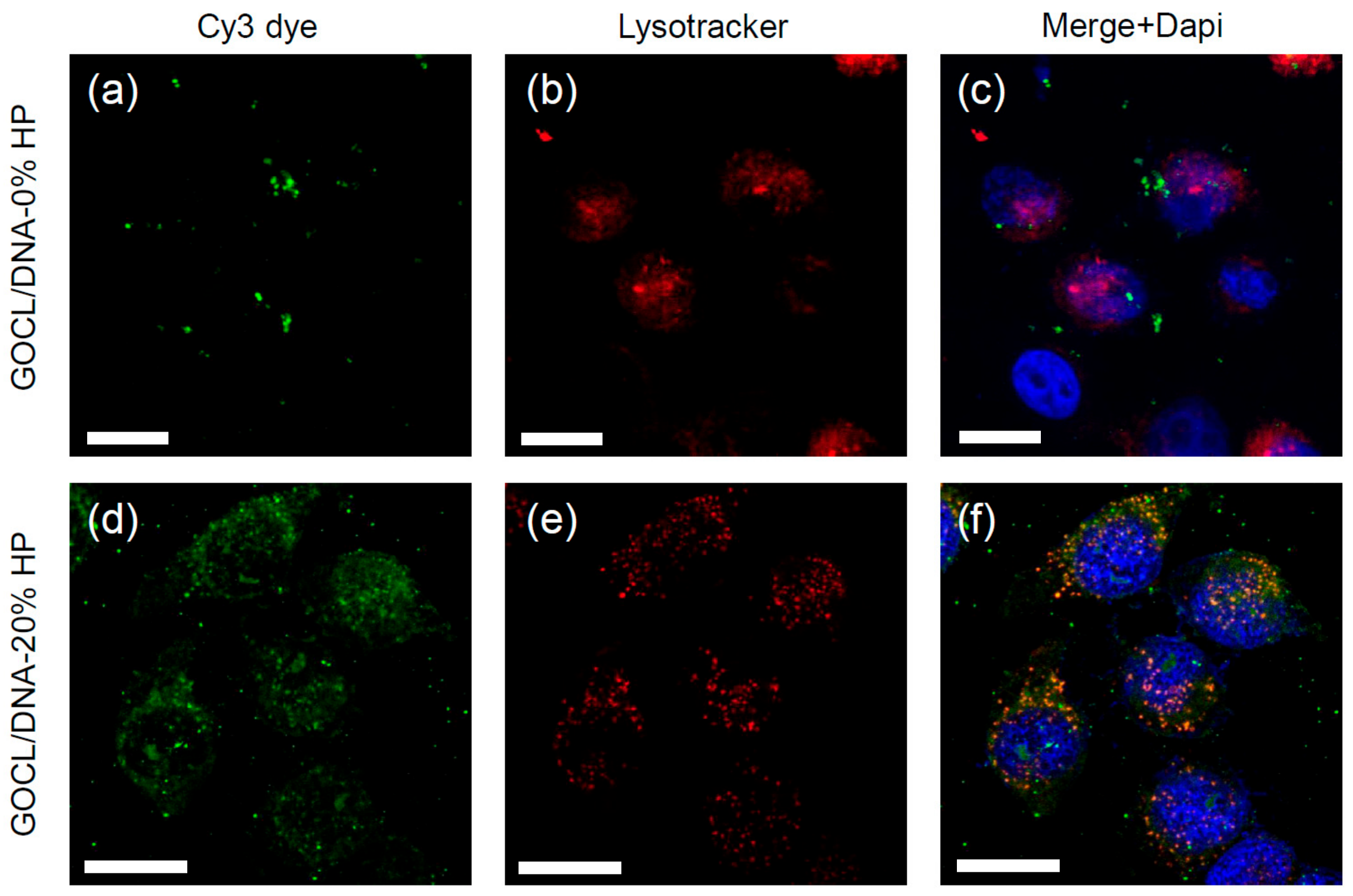
3.5. Intracellular Final Fate of Biocoronated Grapholipoplexes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Di Santo, R.; Digiacomo, L.; Palchetti, S.; Palmieri, V.; Perini, G.; Pozzi, D.; Papi, M.; Caracciolo, G. Microfluidic manufacturing of surface-functionalized graphene oxide nanoflakes for gene delivery. Nanoscale 2019, 11, 2733–2741. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Chen, W.; Liu, F.; Liu, J.; Zhang, T.; Chen, W. Aggregation morphology is a key factor determining protein adsorption on graphene oxide and reduced graphene oxide nanomaterials. Environ. Sci. Nano 2019, 6, 1303–1309. [Google Scholar] [CrossRef]
- Zhang, L.; Xia, J.; Zhao, Q.; Liu, L.; Zhang, Z. Functional graphene oxide as a nanocarrier for controlled loading and targeted delivery of mixed anticancer drugs. small 2010, 6, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Shim, G.; Kim, M.-G.; Park, J.Y.; Oh, Y.-K. Graphene-based nanosheets for delivery of chemotherapeutics and biological drugs. Adv. Drug Deliv. Rev. 2016, 105, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Di Santo, R.; Quagliarini, E.; Palchetti, S.; Pozzi, D.; Palmieri, V.; Perini, G.; Papi, M.; Capriotti, A.L.; Laganà, A.; Caracciolo, G. Microfluidic-generated lipid-graphene oxide nanoparticles for gene delivery. Appl. Phys. Lett. 2019, 114, 233701. [Google Scholar] [CrossRef]
- Cardarelli, F.; Digiacomo, L.; Marchini, C.; Amici, A.; Salomone, F.; Fiume, G.; Rossetta, A.; Gratton, E.; Pozzi, D.; Caracciolo, G. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci. Rep. 2016, 6, 25879. [Google Scholar] [CrossRef]
- Wei, X.-Q.; Hao, L.-Y.; Shao, X.-R.; Zhang, Q.; Jia, X.-Q.; Zhang, Z.-R.; Lin, Y.-F.; Peng, Q. Insight into the interaction of graphene oxide with serum proteins and the impact of the degree of reduction and concentration. Acs Appl. Mater. Interfaces 2015, 7, 13367–13374. [Google Scholar] [CrossRef]
- Caracciolo, G. Liposome–protein corona in a physiological environment: Challenges and opportunities for targeted delivery of nanomedicines. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 543–557. [Google Scholar] [CrossRef]
- Lesniak, A.; Salvati, A.; Santos-Martinez, M.J.; Radomski, M.W.; Dawson, K.A.; Åberg, C. Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency. J. Am. Chem. Soc. 2013, 135, 1438–1444. [Google Scholar] [CrossRef]
- Caracciolo, G.; Farokhzad, O.C.; Mahmoudi, M. Biological identity of nanoparticles in vivo: Clinical implications of the protein corona. Trends Biotechnol. 2017, 35, 257–264. [Google Scholar] [CrossRef]
- Papi, M.; Palmieri, V.; Palchetti, S.; Pozzi, D.; Digiacomo, L.; Guadagno, E.; del Basso De Caro, M.; Di Domenico, M.; Ricci, S.; Pani, R. Exploitation of nanoparticle-protein interactions for early disease detection. Appl. Phys. Lett. 2019, 114, 163702. [Google Scholar] [CrossRef]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, D.; Caracciolo, G.; Digiacomo, L.; Colapicchioni, V.; Palchetti, S.; Capriotti, A.; Cavaliere, C.; Chiozzi, R.Z.; Puglisi, A.; Laganà, A. The biomolecular corona of nanoparticles in circulating biological media. Nanoscale 2015, 7, 13958–13966. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Pozzi, D.; Capriotti, A.L.; Cavaliere, C.; Foglia, P.; Amenitsch, H.; Laganà, A. Evolution of the protein corona of lipid gene vectors as a function of plasma concentration. Langmuir 2011, 27, 15048–15053. [Google Scholar] [CrossRef]
- Digiacomo, L.; Giulimondi, F.; Mahmoudi, M.; Caracciolo, G. Effect of molecular crowding on the biological identity of liposomes: An overlooked factor at the bio-nano interface. Nanoscale Adv. 2019. [Google Scholar] [CrossRef]
- Lesniak, A.; Fenaroli, F.; Monopoli, M.P.; Åberg, C.; Dawson, K.A.; Salvati, A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. Acs Nano 2012, 6, 5845–5857. [Google Scholar] [CrossRef]
- Gräfe, C.; Weidner, A.; vd Lühe, M.; Bergemann, C.; Schacher, F.H.; Clement, J.H.; Dutz, S. Intentional formation of a protein corona on nanoparticles: Serum concentration affects protein corona mass, surface charge, and nanoparticle–cell interaction. Int. J. Biochem. Cell Biol. 2016, 75, 196–202. [Google Scholar] [CrossRef]
- Ge, C.; Tian, J.; Zhao, Y.; Chen, C.; Zhou, R.; Chai, Z. Towards understanding of nanoparticle–protein corona. Arch. Toxicol. 2015, 89, 519–539. [Google Scholar] [CrossRef]
- La Barbera, G.; Capriotti, A.L.; Caracciolo, G.; Cavaliere, C.; Cerrato, A.; Montone, C.M.; Piovesana, S.; Pozzi, D.; Quagliarini, E.; Laganà, A. A comprehensive analysis of liposomal biomolecular corona upon human plasma incubation: The evolution towards the lipid corona. Talanta 2019, 120487. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical−chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef]
- Berardi, A.; Baldelli Bombelli, F. Oral Delivery of Nanoparticles-let’s not Forget About the Protein Corona; Taylor & Francis: Abingdon, UK, 2019. [Google Scholar]
- Raesch, S.S.; Tenzer, S.; Storck, W.; Rurainski, A.; Selzer, D.; Ruge, C.A.; Perez-Gil, J.; Schaefer, U.F.; Lehr, C.-M. Proteomic and lipidomic analysis of nanoparticle corona upon contact with lung surfactant reveals differences in protein, but not lipid composition. Acs Nano 2015, 9, 11872–11885. [Google Scholar] [CrossRef] [PubMed]
- Pecora, R. Dynamic Light Scattering Measurement of Nanometer Particles in Liquids. J. Nanoparticle Res. 2000, 2, 123–131. [Google Scholar] [CrossRef]
- Carpenter, D.K. Dynamic Light Scattering with Applications to Chemistry, Biology, and Physics (Berne, Bruce J.; Pecora, Robert). J. Chem. Educ. 1977, 54, A430. [Google Scholar] [CrossRef]
- Hunter, R. Zeta Potential in Colloid Science: Principles and Applications; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Palchetti, S.; Caputo, D.; Digiacomo, L.; Capriotti, A.L.; Coppola, R.; Pozzi, D.; Caracciolo, G. Protein Corona Fingerprints of Liposomes: New Opportunities for Targeted Drug Delivery and Early Detection in Pancreatic Cancer. Pharmaceutics 2019, 11, 31. [Google Scholar] [CrossRef]
- Lang, F. Mechanisms and significance of cell volume regulation. J. Am. Coll. Nutr. 2007, 26, 613S–623S. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, D.; Caracciolo, G.; Caminiti, R.; De Sanctis, S.C.; Amenitsch, H.; Marchini, C.; Montani, M.; Amici, A. Toward the Rational Design of Lipid Gene Vectors: Shape Coupling between Lipoplex and Anionic Cellular Lipids Controls the Phase Evolution of Lipoplexes and the Efficiency of DNA Release. ACS Appl. Mater. Interfaces 2009, 1, 2237–2249. [Google Scholar] [CrossRef]
- Caracciolo, G.; Amenitsch, H. Cationic liposome/DNA complexes: From structure to interactions with cellular membranes. Eur. Biophys. J. 2012, 41, 815–829. [Google Scholar] [CrossRef]
- Zuzzi, S.; Cametti, C.; Onori, G. Polyion-induced aggregation of lipidic-coated solid polystyrene spheres: The many facets of complex formation in low-density colloidal suspensions. Langmuir 2008, 24, 6044–6049. [Google Scholar] [CrossRef]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Patri, A.K.; Zheng, J.; Clogston, J.D.; Ayub, N.; Aggarwal, P.; Neun, B.W.; Hall, J.B.; McNeil, S.E. Interaction of colloidal gold nanoparticles with human blood: Effects on particle size and analysis of plasma protein binding profiles. Nanomed. Nanotechnol. Biol. Med. 2009, 5, 106–117. [Google Scholar] [CrossRef]
- Amenitsch, H.; Caracciolo, G.; Foglia, P.; Fuscoletti, V.; Giansanti, P.; Marianecci, C.; Pozzi, D.; Laganà, A. Existence of hybrid structures in cationic liposome/DNA complexes revealed by their interaction with plasma proteins. Colloids Surf. B Biointerfaces 2011, 82, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Ruenraroengsak, P.; Novak, P.; Berhanu, D.; Thorley, A.J.; Valsami-Jones, E.; Gorelik, J.; Korchev, Y.E.; Tetley, T.D. Respiratory epithelial cytotoxicity and membrane damage (holes) caused by amine-modified nanoparticles. Nanotoxicology 2012, 6, 94–108. [Google Scholar] [CrossRef]
- Molinaro, R.; Wolfram, J.; Federico, C.; Cilurzo, F.; Di Marzio, L.; Ventura, C.A.; Carafa, M.; Celia, C.; Fresta, M. Polyethylenimine and chitosan carriers for the delivery of RNA interference effectors. Expert Opin. Drug Deliv. 2013, 10, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.; Kang, S.-G.; Tian, X.; Garate, J.A.; Zhao, L.; Ge, C.; Zhou, R. Protein corona mitigates the cytotoxicity of graphene oxide by reducing its physical interaction with cell membrane. Nanoscale 2015, 7, 15214–15224. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Peng, C.; Lv, M.; Li, X.; Zhang, Y.; Chen, N.; Fan, C.; Huang, Q. Protein corona-mediated mitigation of cytotoxicity of graphene oxide. Acs Nano 2011, 5, 3693–3700. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Molinaro, R.; Parodi, A.; Toledano Furman, N.E.; Salvatore, F.; Tasciotti, E. The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine 2016, 11, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 2009, 8, 543. [Google Scholar] [CrossRef]
- Caracciolo, G.; Palchetti, S.; Colapicchioni, V.; Digiacomo, L.; Pozzi, D.; Capriotti, A.L.; La Barbera, G.; Laganà, A. Stealth Effect of Biomolecular Corona on Nanoparticle Uptake by Immune Cells. Langmuir 2015, 31, 10764–10773. [Google Scholar] [CrossRef]
- Von Karstedt, S.; Montinaro, A.; Walczak, H. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Rev. Cancer 2017, 17, 352. [Google Scholar] [CrossRef]
- Bigdeli, A.; Palchetti, S.; Pozzi, D.; Hormozi-Nezhad, M.R.; Baldelli Bombelli, F.; Caracciolo, G.; Mahmoudi, M. Exploring Cellular Interactions of Liposomes Using Protein Corona Fingerprints and Physicochemical Properties. Acs Nano 2016, 10, 3723–3737. [Google Scholar] [CrossRef]
- Palchetti, S.; Digiacomo, L.; Pozzi, D.; Peruzzi, G.; Micarelli, E.; Mahmoudi, M.; Caracciolo, G. Nanoparticles-cell association predicted by protein corona fingerprints. Nanoscale 2016, 8, 12755–12763. [Google Scholar] [CrossRef] [PubMed]
- Kruyt, F. Multiple interactions between cancer cells and the tumor microenvironment modulate TRAIL signaling: Implications for TRAIL receptor targeted therapy. Front. Immunol. 2019, 10, 1530. [Google Scholar]
- Ye, D.; Raghnaill, M.N.; Bramini, M.; Mahon, E.; Åberg, C.; Salvati, A.; Dawson, K.A. Nanoparticle accumulation and transcytosis in brain endothelial cell layers. Nanoscale 2013. [Google Scholar] [CrossRef]
- Bramini, M.; Ye, D.; Hallerbach, A.; Nic Raghnaill, M.; Salvati, A.; Åberg, C.; Dawson, K.A. Imaging approach to mechanistic study of nanoparticle interactions with the blood–brain barrier. Acs Nano 2014, 8, 4304–4312. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, F.; Garry, D.; Monopoli, M.P.; Salvati, A.; Dawson, K.A. The intracellular destiny of the protein corona: A study on its cellular internalization and evolution. Acs Nano 2016, 10, 10471–10479. [Google Scholar] [CrossRef]
- Rehman, Z.u.; Hoekstra, D.; Zuhorn, I.S. Mechanism of polyplex-and lipoplex-mediated delivery of nucleic acids: Real-time visualization of transient membrane destabilization without endosomal lysis. Acs Nano 2013, 7, 3767–3777. [Google Scholar] [CrossRef]
- Pozzi, D.; Marchini, C.; Cardarelli, F.; Salomone, F.; Coppola, S.; Montani, M.; Zabaleta, M.E.; Digman, M.A.; Gratton, E.; Colapicchioni, V.; et al. Mechanistic evaluation of the transfection barriers involved in lipid-mediated gene delivery: Interplay between nanostructure and composition. Biochim. Et Biophys. Acta Biomembr. 2014, 1838, 957–967. [Google Scholar] [CrossRef]
- Pozzi, D.; Marchini, C.; Cardarelli, F.; Rossetta, A.; Colapicchioni, V.; Amici, A.; Montani, M.; Motta, S.; Brocca, P.; Cantù, L.; et al. Mechanistic understanding of gene delivery mediated by highly efficient multicomponent envelope-type nanoparticle systems. Mol. Pharm. 2013, 10, 4654–4665. [Google Scholar] [CrossRef]
- Giulimondi, F.; Digiacomo, L.; Pozzi, D.; Palchetti, S.; Vulpis, E.; Capriotti, A.L.; Chiozzi, R.Z.; Laganà, A.; Amenitsch, H.; Masuelli, L. Interplay of protein corona and immune cells controls blood residency of liposomes. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Capriotti, A.L.; Caracciolo, G.; Cavaliere, C.; Foglia, P.; Pozzi, D.; Samperi, R.; Laganà, A. Do plasma proteins distinguish between liposomes of varying charge density? J. Proteom. 2012, 75, 1924–1932. [Google Scholar] [CrossRef]
- Caracciolo, G.; Cardarelli, F.; Pozzi, D.; Salomone, F.; Maccari, G.; Bardi, G.; Capriotti, A.L.; Cavaliere, C.; Papi, M.; Laganà, A. Selective targeting capability acquired with a protein corona adsorbed on the surface of 1,2-dioleoyl-3-trimethylammonium propane/dna nanoparticles. Acs Appl. Mater. Interfaces 2013, 5, 13171–13179. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Müller, L.K.; Kokkinopoulou, M.; Lieberwirth, I.; Morsbach, S.; Landfester, K.; Mailänder, V. Exploiting the biomolecular corona: Pre-coating of nanoparticles enables controlled cellular interactions. Nanoscale 2018, 10, 10731–10739. [Google Scholar] [CrossRef]
- Digiacomo, L.; Cardarelli, F.; Pozzi, D.; Palchetti, S.; Digman, M.; Gratton, E.; Capriotti, A.; Mahmoudi, M.; Caracciolo, G. An apolipoprotein-enriched biomolecular corona switches the cellular uptake mechanism and trafficking pathway of lipid nanoparticles. Nanoscale 2017, 9, 17254–17262. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quagliarini, E.; Di Santo, R.; Palchetti, S.; Ferri, G.; Cardarelli, F.; Pozzi, D.; Caracciolo, G. Effect of Protein Corona on The Transfection Efficiency of Lipid-Coated Graphene Oxide-Based Cell Transfection Reagents. Pharmaceutics 2020, 12, 113. https://doi.org/10.3390/pharmaceutics12020113
Quagliarini E, Di Santo R, Palchetti S, Ferri G, Cardarelli F, Pozzi D, Caracciolo G. Effect of Protein Corona on The Transfection Efficiency of Lipid-Coated Graphene Oxide-Based Cell Transfection Reagents. Pharmaceutics. 2020; 12(2):113. https://doi.org/10.3390/pharmaceutics12020113
Chicago/Turabian StyleQuagliarini, Erica, Riccardo Di Santo, Sara Palchetti, Gianmarco Ferri, Francesco Cardarelli, Daniela Pozzi, and Giulio Caracciolo. 2020. "Effect of Protein Corona on The Transfection Efficiency of Lipid-Coated Graphene Oxide-Based Cell Transfection Reagents" Pharmaceutics 12, no. 2: 113. https://doi.org/10.3390/pharmaceutics12020113
APA StyleQuagliarini, E., Di Santo, R., Palchetti, S., Ferri, G., Cardarelli, F., Pozzi, D., & Caracciolo, G. (2020). Effect of Protein Corona on The Transfection Efficiency of Lipid-Coated Graphene Oxide-Based Cell Transfection Reagents. Pharmaceutics, 12(2), 113. https://doi.org/10.3390/pharmaceutics12020113